How to use this server
Step 1: Upload Busco data
• Upload file- Upload your multi-FASTA file.
• Custimize your input by choosing outgroup, lineage, mode and name of the project.
• Submit the job by clicking on Submit button.

Step 2:
• If you are logged into the app : The projects will be stored automatically into your Account page.
• Else : You can find your projects using the job ID or by checking your mailbox.
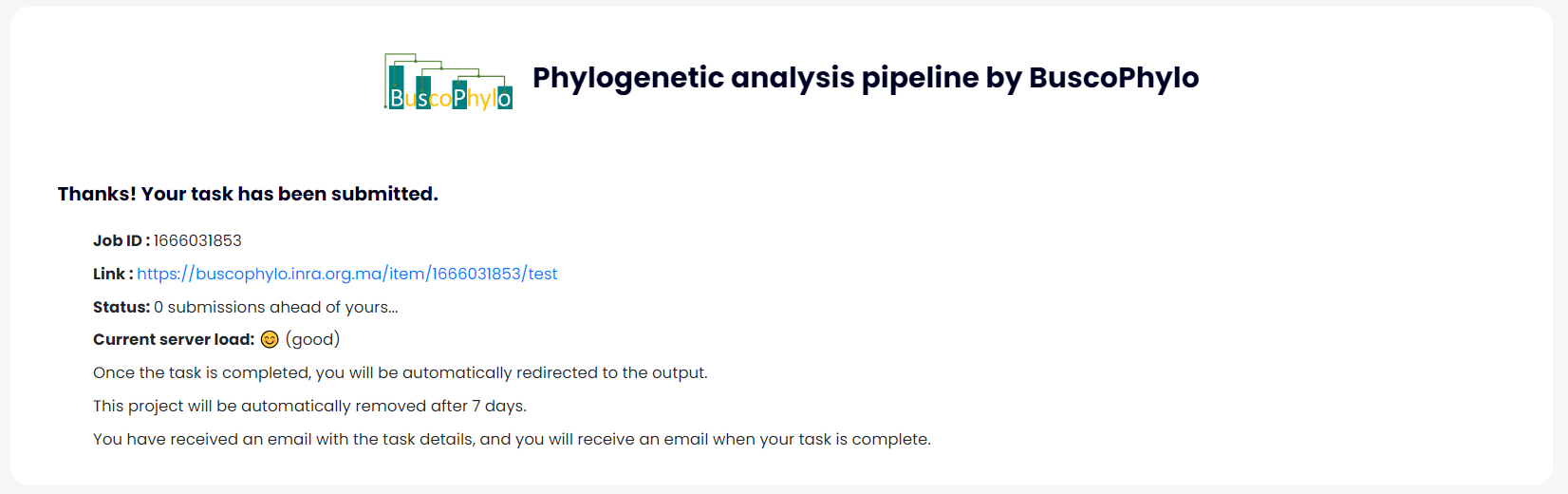
Step 3: Results
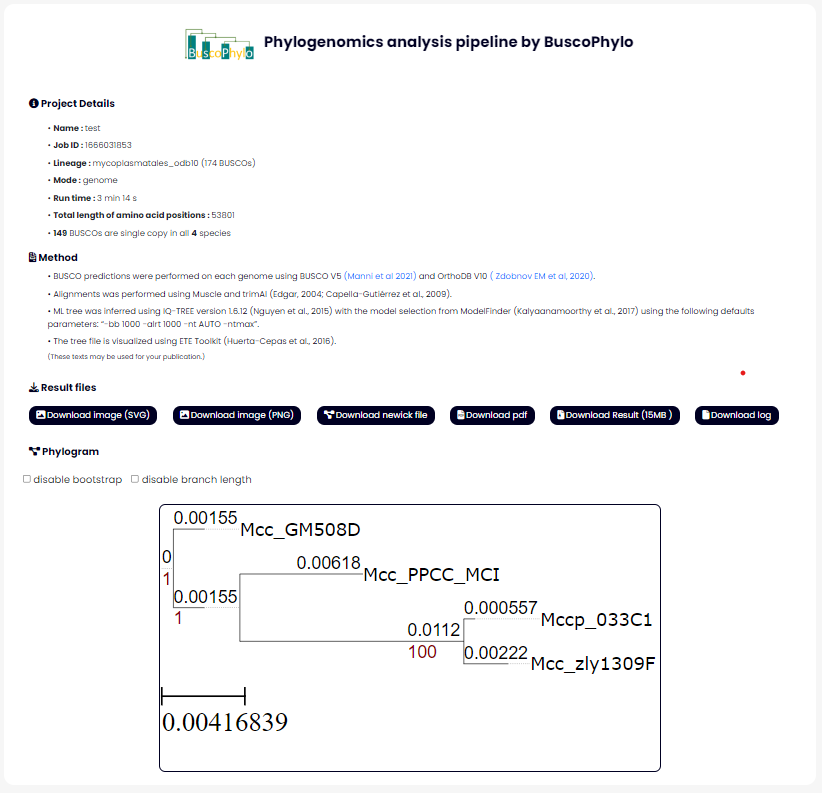
• Once the file upload has been completed, the following results page will be shown; you can download the result as a PNG or PDF or you can download the whole PROJECT FOLDER as a ZIP using the buttons at the bottom of the result section.
How to use the app in local server
Thanks to the advances in DNA sequencing technolgies, thousands of genome sequences of living organisms are being released in the public databases every day. BuscoPhylo has been implemented to provide a fully automated and complete pipeline to quickly perform BUSCO-based phylogenomic analysis starting from assembled genome sequences as inputs. The BuscoPhylo is a free, on-line and user-friendly webserver accepting genome sequences in FASTA format as inputs and enabling to the user to export the tree ready for publication and all the results of the steps included in the pipeline for downstream analyses.
Installation
- Download the application and extract the zipped file in your web server (xampp, ampps, WAMP, online server …)
# Download the BuscoPhylo source code
git clone https://github.com/alaesahbou/BuscoPhylo.git
# Move the the BuscoPhylo dir to your your server (exmaple here lampp)
mv BuscoPhylo/ /opt/lampp/htdocs/
- Setup the server
# Give BuscoPhylo root privileges using the following command
sudo chown -R daemon /BuscoPhylo_directory
sudo visudo
# insert th following lines after User privilege specification comment
# for xampp replace USER by daemon and for ampps replace USER by ampps
[USER] ALL=(ALL:ALL) ALL
[USER] ALL=(ALL) NOPASSWD: ALL
# open the file with a text manger
vim /etc/php5/cli/php.ini
# change these lines:
max_file_uploads=5000
upload_max_filesize=8000M
post_max_size=8000M
- Open BuscoPhylo tool via localhost on you browser (http://localhost/BuscoPhylo/)
- Connect BuscoPhylo to mysql
Enter your login and password of your mysql server

Requirements
Submitting a project
- Input Requirements
- At least 4 Genome sequence files in FASTA format (.fa,.fsa,.fasta,.fna are supported)
- File names will be used as leaf labels
- We recommand this format
Genus_species_strain.fasta
GUI input portal

Output files
- outBusco directory containing Busco runs
- out directory containing the ML tree, logs, BSCg as proteins and alignments
- pdf png svg files for phylogenomic tree
License
BuscoPhylo is licensed under the MIT License.
Citations
If BuscoPhylo helped with the analysis of your data, please do not forget to cite:
Phylogenomics analyses were conducted in BuscoPhylo (Sahbou et al., 2022).
BUSCO predictions were performed on each genome using BUSCO V5 (Manni et al 2021) and OrthoDB V10 ( Zdobnov EM et al, 2020).
Muscle and trimAl (Edgar, 2004; Capella-Gutiérrez et al., 2009).
ML tree was inferred using IQ-TREE version 1.6.12 (Nguyen et al., 2015) with the model selection from ModelFinder (Kalyaanamoorthy et al., 2017) using the following defaults parameters: “-bb 1000 -alrt 1000 -nt AUTO -ntmax”.
The tree file is visualized using ETE Toolkit (Huerta-Cepas et al., 2016).





